Clustering Algorithm Demo
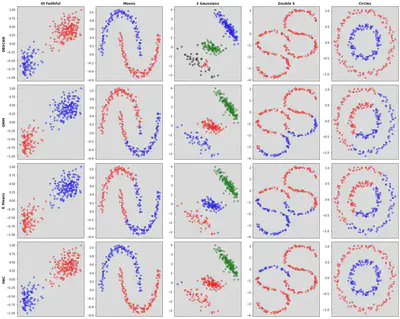
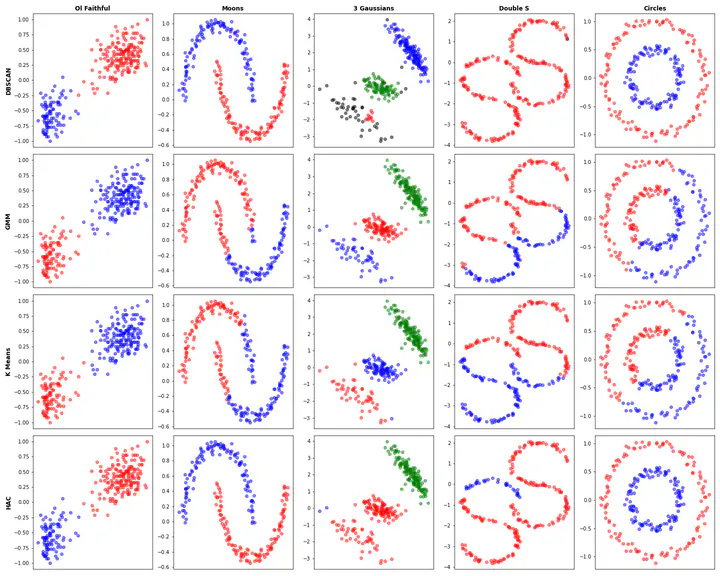 Overall comparison of these 4 clustering algorithms on 5 datasets.
Overall comparison of these 4 clustering algorithms on 5 datasets.Imports and Common Functions
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import minimize
from scipy.stats import t, norm, beta, multivariate_normal
from scipy.spatial import distance
from sklearn import datasets
import copy
import matplotlib.colors as mcolors
def euclidDist(x1,x2):
return np.sqrt(np.sum((x1-x2)**2))
colors = ['k','r','b','g','c','m','y']
colors.extend(set(mcolors.CSS4_COLORS))
Gaussian Mixture Model Optimization with Expectation Maximization
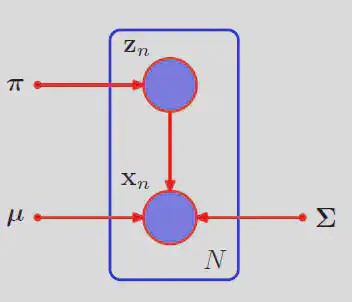
The GMM model works by assuming the data was genrated by a set of
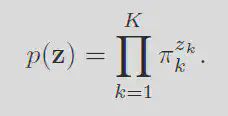
Every position
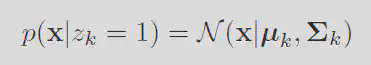
Then by the rules of conditional probability, we can sum over every configuration of
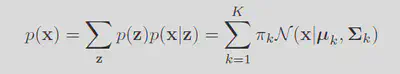
This form of a likelihood implies a sampling struture. Each data point
With the likelihood sorted, simply we just maximize the probability of observing the data. Note the fact that maximizing the regular likelihood is equivilent to maximizing the log of it. Giving the log probability as:
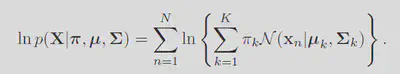
Taking the derivative of this function with respect to the means
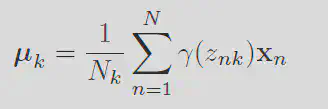
Define the responsibility
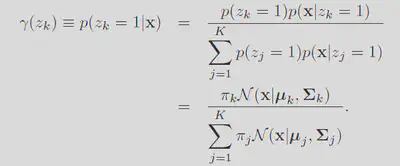
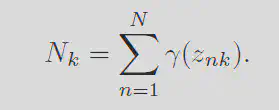
Each of these terms can be interpereted with respect to the orginal problem. The responsibility is like the
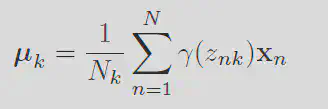
The variance can be solved similarly.
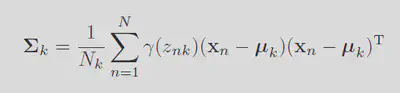
For the
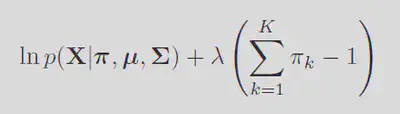
Then differentiating gives:
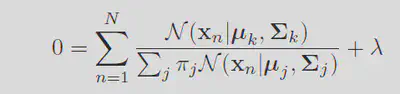
Because we know that the sum of
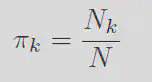
This may seem great, however note the fact that every term complexly depends on the responsibility
def γ_set(X,μ_arr,Σ_arr,π_arr):
K = len(μ_arr)
# Create an array to store each \pi_k*N(X|\mu_k,\Sigma_k) such that it is indexed by the cluster index k
mvnArray = np.zeros((K,len(X[0])))
for k in range(K):
mvn = multivariate_normal(μ_arr[k],Σ_arr[k])
mvnArray[k] = mvn.pdf(X)*π_arr[k]
# Compute the sum of each and then use that to normalize them
Z = np.sum(mvnArray,0)
for k in range(K):
mvnArray[k] /= Z
return mvnArray
def LogLH(X,μ_arr,Σ_arr,π_arr):
LH = 0
for i in range(len(π_arr)):
mvnSub = multivariate_normal(μ_arr[i],Σ_arr[i])
LH += mvnSub.pdf(X)*π_arr[i]
return np.sum(np.log(LH))
# Compute the results of GMM algorithm
# X = a 1 by N by D array such that N is the number of samples and D corresponds to the dimensonality of the data
# μ_init = The initial mean locations, K by D. Where K is the number of clusters
# Σ_init = Initial Variance Matrix, K by D by D.
# π_init = Initial probability vector of belonging to cluster k. It must sum to 1. K by 1.
# returnPath = True if you want to see the path π,μ, and Σ takes along their optimization
def GMM(X, μ_init, Σ_init, π_init,returnPath= False):
K = len(μ_init)
#Initialize fields and orgnize data.
μ = μ_init
Σ = Σ_init
π = π_init
# Grade the convergence based on the change in the log LH.
# Once the chage in the LLH changes by less than this value the algorithm will stop.
tolerance = 1e-20
ΔLLH = 9999999 # Init delta to be super large
LLH = 0
MAX_ITERATIONS = 100
iteration_count = 0
# Store evaluated parameters on our journey to the minimum
π_path = np.empty((MAX_ITERATIONS+1,K))
μ_path = np.empty((MAX_ITERATIONS+1,K,2))
Σ_path = np.empty((MAX_ITERATIONS+1,K,2,2))
μ_path[0] = μ_init
Σ_path[0] = Σ_init
π_path[0] = π_init
# Run until convergence of LLH or until we hit the max iterations.
while iteration_count < MAX_ITERATIONS and ΔLLH > tolerance:
# EXPECTATION STEP
# Compute the current responsibility for each class
Γ = γ_set(X,μ,Σ,π) # (K,N) for ol faith (2,272)
N = np.sum(Γ,1) # (K) for ol faith (2,)
# MAXIMIZATION STEP
μ_new = (Γ @ X[0]) # \sum_N \gamma_kn*\X_n (D,K) for ol faith (2,2)
for k in range(len(N)):
μ_new[k] /= N[k]
Σ_new = np.zeros((len(Σ),len(μ[0]),len(μ[0])))
for k in range(len(Σ)):
diff = (X[0] - μ[k])
for d in range(len(X[0][0])):
diff[:,d] *= Γ[k]
Σ_new[k] = (1/N[k])*((X[0] - μ[k]).T @ diff)
π_new = N/len(X[0])
LLH_new = LogLH(X,μ,Σ,π)
# Calculate Tolerance
ΔLLH = abs(LLH_new - LLH)
LLH = LLH_new
iteration_count += 1
# Save Maximized Values
μ = μ_new
Σ = Σ_new
π = π_new
μ_path[iteration_count] = μ
Σ_path[iteration_count] = Σ
π_path[iteration_count] = π
# Creating a place to store the classification of the datum
classification = []
# In order to aid in plotting, ignore the soft classification and just assign the highest probability
# In general this is not right but what can you do
for point in X[0]:
maxP = 0
index = 0
for k in range(len(π)):
mvn = multivariate_normal(μ[k],Σ[k])
p_k = mvn.pdf(np.array(point))
if p_k > maxP:
maxP = mvn.pdf(np.array(point))
index = k
classification.append(index)
if returnPath:
print("Steps to convergence:", iteration_count)
return classification, μ_path, Σ_path, π_path
return classification
TEST GMM
from matplotlib.pyplot import figure
X_1 = np.random.multivariate_normal(np.array([-5,-5]),datasets.make_spd_matrix(2),100)
X_2 = np.random.multivariate_normal(np.array([5,5]),datasets.make_spd_matrix(2)/2,100)
X_3 = np.random.multivariate_normal(np.array([0,2]),datasets.make_spd_matrix(2)/3,100)
X = np.array([np.concatenate((X_1,X_2,X_3))])
μ_init = np.array([[0,0] for i in range(3)])
μ_init[0] = np.array([3,-6])
μ_init[1] = np.array([0,0])
μ_init[2] = np.array([5,4])
Σ_init= np.array([np.identity(2)*0.1 for i in range(3)])
π_init = np.array([0.5,0.25,0.25])
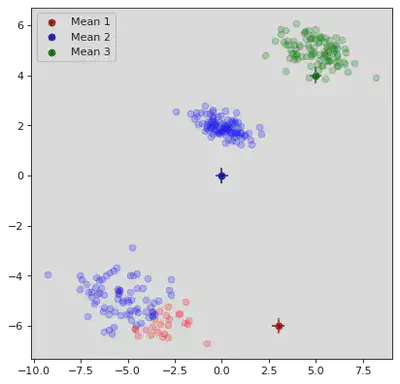
classification, μ_path, Σ_path, π_path = GMM(X, μ_init, Σ_init, π_init,True)
Steps to convergence: 13
for step in range(0,13,2):
figure(figsize=(6, 6), dpi=80)
# Plot Datum
# Abuse the soft classification.
for point in X[0]:
maxP = 0
index = 0
for k in range(len(π_init)):
mvn = multivariate_normal(μ_path[step][k],Σ_path[step][k])
p_k = mvn.pdf(np.array(point))
if p_k > maxP:
maxP = mvn.pdf(np.array(point))
index = k
plt.scatter(point[0],point[1],color=colors[index + 1],alpha=0.25)
# Plot Means:
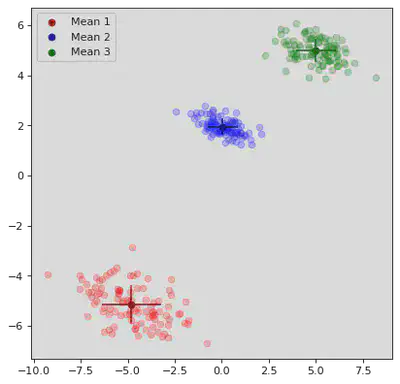
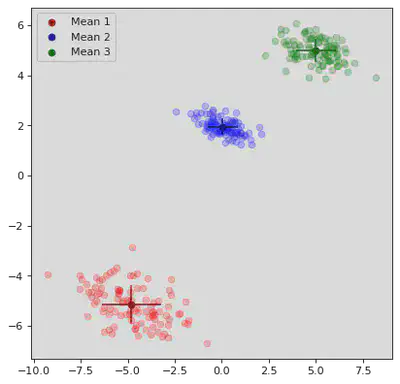
plt.scatter(μ_path[step][0][0],μ_path[step][0][1],color='darkred',label="Mean 1",marker="o")
plt.scatter(μ_path[step][1][0],μ_path[step][1][1],color='darkblue',label="Mean 2",marker="o")
plt.scatter(μ_path[step][2][0],μ_path[step][2][1],color='darkgreen',label="Mean 3",marker="o")
# Plot Means:
xstd = np.sqrt(Σ_path[step][0][0][0])
ystd = np.sqrt(Σ_path[step][0][1][1])
plt.vlines(μ_path[step][0][0],μ_path[step][0][1] - ystd,μ_path[step][0][1] + ystd,color='darkred')
plt.hlines(μ_path[step][0][1],μ_path[step][0][0] - xstd,μ_path[step][0][0] + xstd,color='darkred')
xstd = np.sqrt(Σ_path[step][1][0][0])
ystd = np.sqrt(Σ_path[step][1][1][1])
plt.vlines(μ_path[step][1][0],μ_path[step][1][1] - ystd,μ_path[step][1][1] + ystd,color='darkblue')
plt.hlines(μ_path[step][1][1],μ_path[step][1][0] - xstd,μ_path[step][1][0] + xstd,color='darkblue')
xstd = np.sqrt(Σ_path[step][2][0][0])
ystd = np.sqrt(Σ_path[step][2][1][1])
plt.vlines(μ_path[step][2][0],μ_path[step][2][1] - ystd,μ_path[step][2][1] + ystd,color='darkgreen')
plt.hlines(μ_path[step][2][1],μ_path[step][2][0] - xstd,μ_path[step][2][0] + xstd,color='darkgreen')
plt.legend()
plt.show()
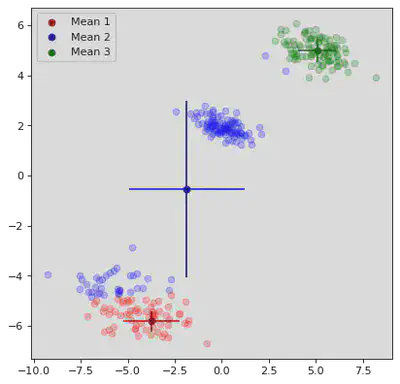
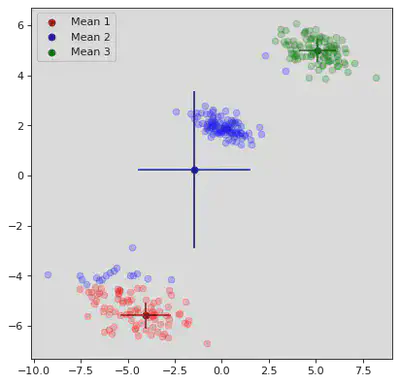
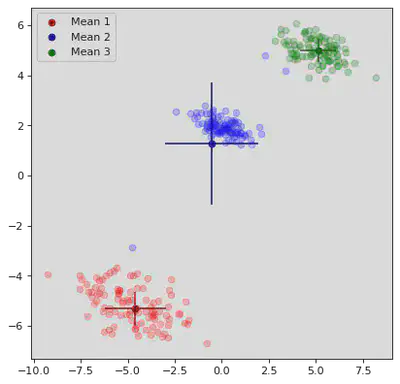
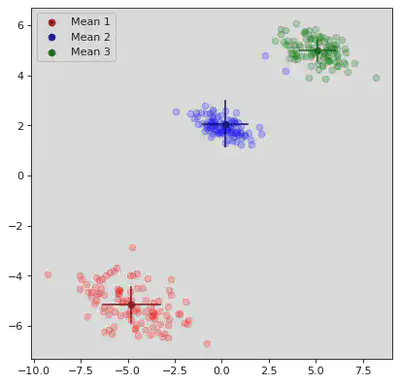
K Means
K means clusterings goal can be summaraized in the definition of the distortion measure
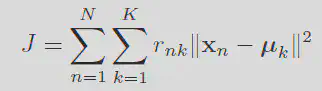
The value
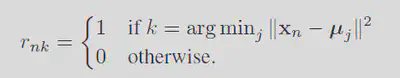
Similarly, by fixing the r values you can minimize the means by differentiation. Setting the deriviative to zero gives:
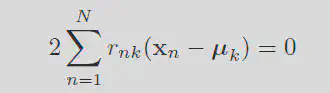
Solving for mu gives:
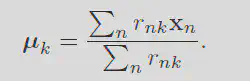
By first minimizing w.r.t. r then w.r.t. mu you can iteratively find the best set of means.
Relation to Gaussian Mixture Models
K means is simply a limiting case of the Gaussain mixture model. Consider the probability of the data given a particular cluster mean and variance
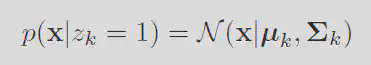
By choosing the variance for every cluster as a fixed parameter
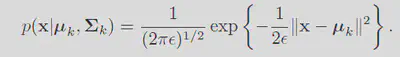
Remebering the responsibilities is defined as:
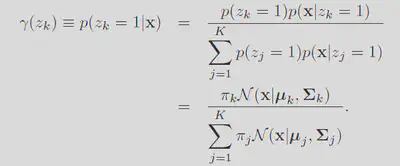
Applying this formula to the current likelihood will then give:
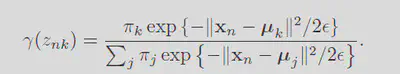
Now imagine taking the limit where
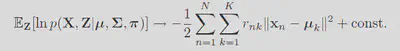
By maximizing the expected log likelihood is the same as minimizing the distortion measure used in K means. GMM is just a generic K means algorithm.
def KMEANS(X,μ_init,returnPath = False):
# Initialize Datas
N = len(X[0])
K = len(μ_init)
μ = μ_init # means, k,1
r = np.empty((N,K))
# Grade the convergence based on the change in the μ.
# Once the chage in μ changes by less than this value the algorithm will stop.
tolerance = 1e-20
Δμ = 9999999 # Init delta to be super large
MAX_ITERATIONS = 100
r_path = np.empty((MAX_ITERATIONS+1,N,K))
μ_path = np.empty((MAX_ITERATIONS+1,K,2))
iteration_count = 0
while iteration_count < MAX_ITERATIONS and Δμ > tolerance:
μ_path[iteration_count] = μ
# EXPECTATION STEP
# Compute best set of indicator variables given means
for n in range(N):
minIndex = 0
minDist = 10000000000000000000 # big numba haha
for k in range(K):
squaredErr = (euclidDist(X[0][n],μ[k]))**2
if (squaredErr < minDist):
minDist = squaredErr
minIndex = k
r[n] = np.zeros(K)
r[n][minIndex] = 1
r_path[iteration_count] = r
# MAXIMIZATION STEP
# Compute best set of means given indicator variables
R = np.sum(r,axis=0)
rT = np.transpose(r)
oldμ = μ
μ = (rT @ X[0])
for k in range(K):
μ[k]/= R[k]
Δμ = np.sum((μ-oldμ)**2)
iteration_count += 1
# Compute the final cluster ownership to return to the user
classification = []
for n in range(N):
minIndex = 0
minDist = 10000000000000000000 # big numba haha
for k in range(K):
squaredErr = (euclidDist(X[0][n],μ[k]))**2
if (squaredErr < minDist):
minDist = squaredErr
minIndex = k
classification.append(minIndex)
if returnPath:
print("Steps to convergence:", iteration_count)
return classification, μ_path, r_path
return classification
TEST KMEANS
from matplotlib.pyplot import figure
X_1 = np.random.multivariate_normal(np.array([-5,-5]),datasets.make_spd_matrix(2),100)
X_2 = np.random.multivariate_normal(np.array([5,5]),datasets.make_spd_matrix(2)/2,100)
X_3 = np.random.multivariate_normal(np.array([0,2]),datasets.make_spd_matrix(2)/3,100)
X = np.array([np.concatenate((X_1,X_2,X_3))])
μ_init = np.array([[0,0] for i in range(3)])
μ_init[0] = np.array([3,-6])
μ_init[1] = np.array([0,0])
μ_init[2] = np.array([5,4])
classification, μ_path, r_path = KMEANS(X,μ_init,True)
Steps to convergence: 4
for step in range(0,4):
figure(figsize=(6, 6), dpi=80)
# Plot Datum:
for i in range(len(X[0])):
plt.scatter(X[0][i][0],X[0][i][1],color=colors[np.where(r_path[step][i] == 1)[0][0]+1],alpha=0.25)
# Plot Means:
plt.scatter(μ_path[step][0][0],μ_path[step][0][1],color='darkred',label="Mean 1",marker="$\mu$",s=50)
plt.scatter(μ_path[step][1][0],μ_path[step][1][1],color='darkblue',label="Mean 2",marker="$\mu$",s=50)
plt.scatter(μ_path[step][2][0],μ_path[step][2][1],color='darkgreen',label="Mean 3",marker="$\mu$",s=50)
plt.legend()
plt.show()
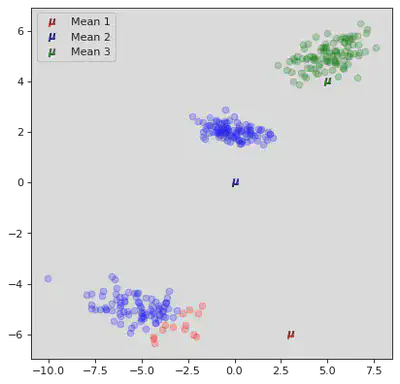
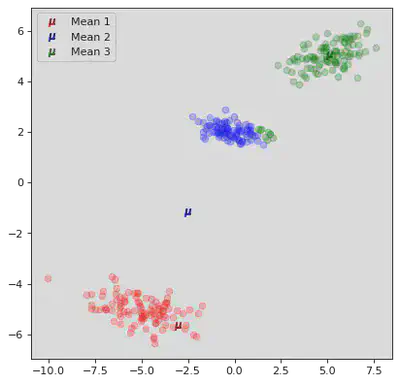
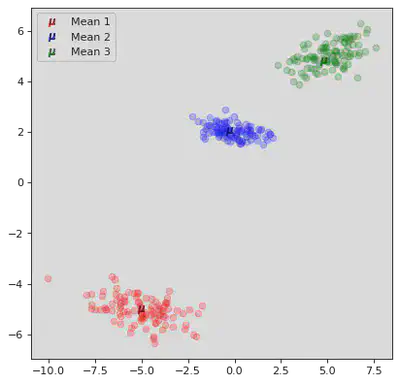
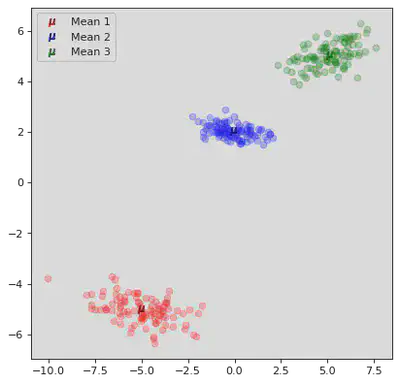
DB Scan
DB scan works by chosing an effective radius
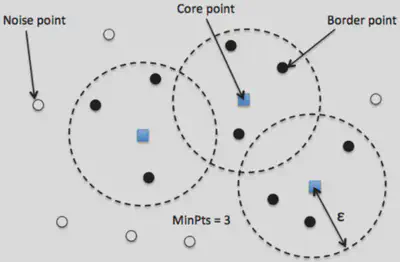
These classes implicitly define a set of classifications. If two core points are within each others epsilon radius then they are the same class. All the border points belong to the class of the core point they belong to. Then the noise points are left as noise. This one is a lot simpler than the Gaussian one haha!(When I wrote that last sentence I did not yet code it. I was so wrong lol)
def getNeighborList(point,radius,dist,data):
neighList = []
for neighPoint in data:
if (dist(point,neighPoint[1]) < radius):
neighList.append(neighPoint[0])
return neighList
def DBSCAN(X,ϵ,minNeigh,dist):
data = []
index = 0
for point in X[0]:
data.append((index,point))
index += 1
# 0 <=> noise k ϵ {1,2,3..} <=> cluster k
classification = dict()
k = 0
# Iterate over each point in the data
for i in range(len(data)):
# Check if it has been classified
if not(i in classification):
#If not, get its neighborlist
N = getNeighborList(data[i][1],ϵ,dist,data)
# If the number of neighbors is less than the minimum classify it as noise
if len(N) < minNeigh:
classification[i] = 0
# If its a core point then expand its neighbors
else:
k += 1
classification[i] = k
S = N
for pointIndex in S:
# If it has been previously classified as noise set it to belonging to group k
if pointIndex in classification:
if classification[pointIndex] == 0:
classification[pointIndex] = k
# Check if it has not been classified
if not(pointIndex in classification):
classification[pointIndex] = k
# Get pointIndex's neighbors
N = getNeighborList(data[pointIndex][1],ϵ,dist,data)
# If pointIndex is a core point, add its neighbors to the processing list
if len(N) >= minNeigh:
S += N
returnList = np.zeros(len(X[0]))
for key, value in classification.items():
returnList[key] = value
return returnList
DBSCAN TEST
X_1 = np.random.multivariate_normal(np.array([-3,-3]),datasets.make_spd_matrix(2),50)
X_2 = np.random.multivariate_normal(np.array([2,2]),datasets.make_spd_matrix(2)/2,150)
X_3 = np.random.multivariate_normal(np.array([0,0]),datasets.make_spd_matrix(2)/3,100)
X = np.array([np.concatenate((X_1,X_2,X_3))])
ϵ = [0.5,0.5,0.25]
MinPts = [10,20,10]
for j in range(3):
figure(figsize=(6, 6), dpi=80)
classification = DBSCAN(X,ϵ[j],MinPts[j],euclidDist)
# Plot Datum:
for i in range(len(X[0])):
plt.scatter(X[0][i][0],X[0][i][1],color=colors[int(classification[i])],alpha=0.5)
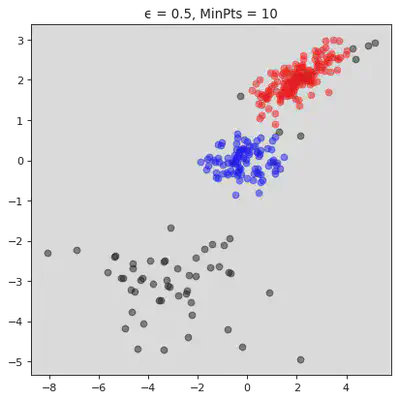
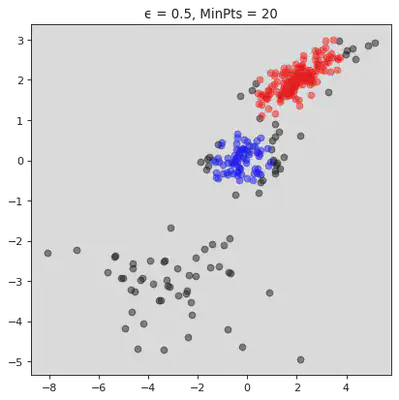
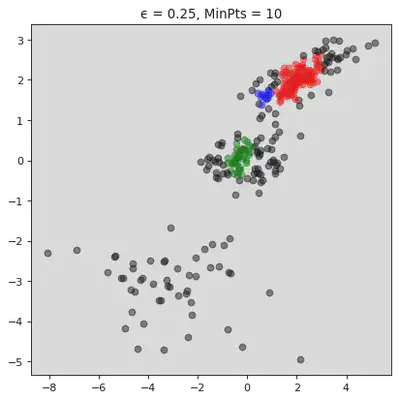
plt.title("ϵ = " + str(ϵ[j]) + ", MinPts = " + str(MinPts[j]))
plt.show()
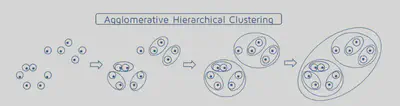
Hierarchical Agglomorative Clustering
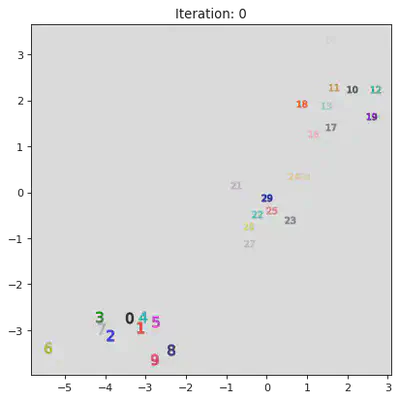
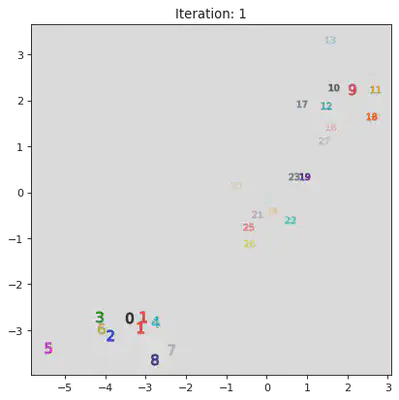
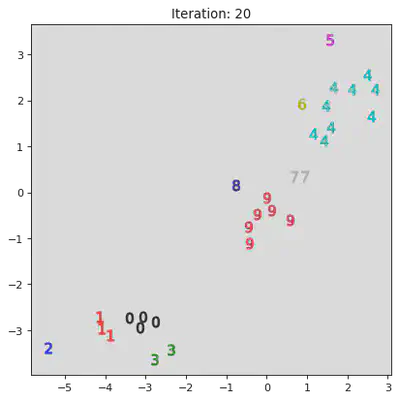
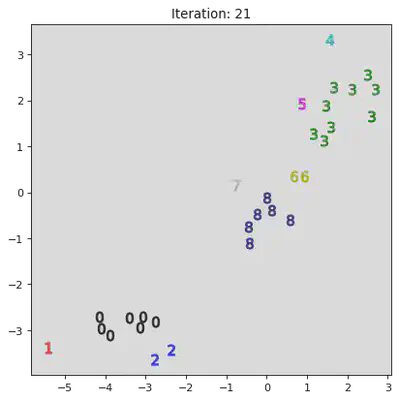
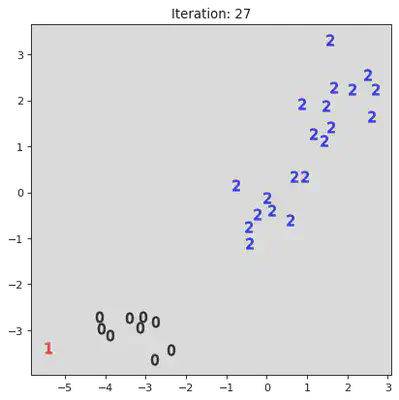
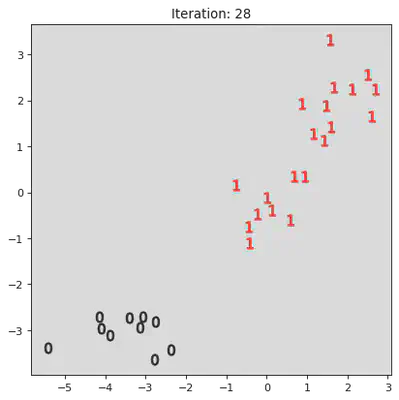
This algorithm works by iteratively combinining groups. Initially the algorithm assigns every datum to its own group. Then it iterativly combines the closest two groups. A pictoral representation of the algorithm can be seen below. Note, not all steps are shown.
The definition of closests heavily influences the algorithm. A few options are shown below.
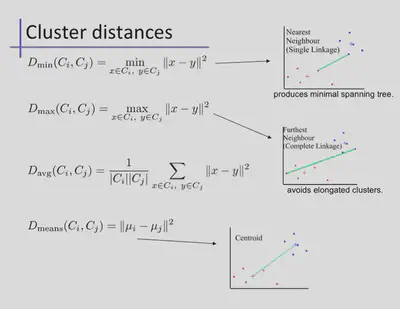
# returns which two groups to merge
def Dmin(classifications,k,RDD):
# Set the default min dist and indices for comparison
minI = 0
minJ = 1
minDist = 100000000000000
# Loop over each group.
for i in range(k):
for j in range(k):
# Ensure we are not in the same group
if not(j == i):
# Set sub min
minDistInGroup = 100000000000000
# Find the minimum distance between elements in each group
for index1 in classifications[i]:
for index2 in classifications[j]:
if(RDD[index1][index2] < minDistInGroup):
minDistInGroup = RDD[index1][index2]
if minDistInGroup < minDist:
minI = i
minJ = j
minDist = minDistInGroup
return minI,minJ
def HAC(X,K,D,returnPath = False):
RDD = distance.cdist(X[0], X[0], 'euclidean')
RDD = RDD + np.identity(len(X[0]))*(max(RDD.flatten())+1)
# k = # of groups
k = len(X[0])
#Init all groups to be on their own
classifications = [[i] for i in range(k)]
classificationsPath = [copy.deepcopy(classifications)]
while k > K:
# Find the two groups to merge
I,J = D(classifications,k,RDD)
lower = min(I,J)
upper = max(I,J)
classifications[lower].extend(classifications[upper])
classifications.pop(upper)
classificationsPath.append(copy.deepcopy(classifications))
k -= 1
classificationsReturn = np.zeros(len(X[0]))
for classIndex in range(K):
for pointIndex in classifications[classIndex]:
classificationsReturn[pointIndex] = classIndex
if returnPath:
return classificationsReturn, classificationsPath
return classificationsReturn
X_1 = np.random.multivariate_normal(np.array([-3,-3]),datasets.make_spd_matrix(2),10)
X_2 = np.random.multivariate_normal(np.array([2,2]),datasets.make_spd_matrix(2)/2,10)
X_3 = np.random.multivariate_normal(np.array([0,0]),datasets.make_spd_matrix(2)/3,10)
X = np.array([np.concatenate((X_1,X_2,X_3))])
classification,classificationsPath = HAC(X,2,Dmin,True)
stepList = [0,1,20,21,27,28]
for step in stepList:
figure(figsize=(6, 6), dpi=80)
classificationsSub = classificationsPath[step]
K = len(classificationsSub)
for classIndex in range(K):
for pointIndex in classificationsSub[classIndex]:
classification[pointIndex] = classIndex
# Plot Datum:
for i in range(len(X[0])):
plt.scatter(X[0][i][0],X[0][i][1],color=colors[int(classification[i])],alpha=0.8,marker="$"+str(int(classification[i]))+"$",s=100)
plt.title("Iteration: " + str(step))
plt.show()
Overall Tests
Ol Faithful Dataset
data = np.zeros((272,2))
for i in range(272):
file = open("data/faithful.txt")
dataTxt = file.read()[19*i:(i+1)*19].split()
data[i] = [float(dataTxt[0]),float(dataTxt[1])]
minX = min(data.T[0])
maxX = max(data.T[0])
minY = min(data.T[1])
maxY = max(data.T[1])
widthX = maxX - minX
widthY = maxY - minY
X_ = np.array(2*(data.T[0] - minX)/widthX -1)
Y_ = np.array(2*(data.T[1] - minY)/widthY - 1)
N_count = 272
X = np.dstack((X_,Y_))
X = np.array([X[0][:N_count]])
XOF = X
classificationDBOF = DBSCAN(X,0.3,10,euclidDist)
μ_init = np.array([[1,1] for i in range(2)])
μ_init[0][0] = -1
μ_init[1][1] = -1
Σ_init= np.array([np.identity(2)*0.1 for i in range(2)])
π_init = np.array([0.5,0.5])
classificationGMMOF = GMM(X, μ_init, Σ_init, π_init)
classificationKMEANSOF = KMEANS(X,μ_init)
classificationHACOF = HAC(X,2,Dmin)
Moons
X = datasets.make_moons(n_samples=250, noise=.05)
X = np.array([X[0]])
XMOON = X
classificationDBMOON = DBSCAN(X,0.3,10,euclidDist)
μ_init = np.array([[1,1] for i in range(2)])
μ_init[0][0] = -1
μ_init[1][1] = -1
Σ_init= np.array([np.identity(2)*0.1 for i in range(2)])
π_init = np.array([0.5,0.5])
classificationGMMMOON = GMM(X, μ_init, Σ_init, π_init)
classificationKMEANSMOON = KMEANS(X,μ_init)
classificationHACMOON = HAC(X,2,Dmin)
Mixture of Gaussians
X_1 = np.random.multivariate_normal(np.random.uniform(-2,-1,2),datasets.make_spd_matrix(2),50)
X_2 = np.random.multivariate_normal(np.random.uniform(2,2.5,2),datasets.make_spd_matrix(2)/2,150)
X_3 = np.random.multivariate_normal(np.random.uniform(0,0,2),datasets.make_spd_matrix(2)/3,100)
X = np.array([np.concatenate((X_1,X_2,X_3))])
XGAUSS = X
classificationDBGAUSS = DBSCAN(X,0.5,10,euclidDist)
μ_init = np.array([[1,1] for i in range(3)])
μ_init[0][0] = -1
μ_init[1][1] = -1
Σ_init= np.array([np.identity(2)*0.1 for i in range(3)])
π_init = np.array([0.5,0.25,0.25])
classificationGMMGAUSS = GMM(X, μ_init, Σ_init, π_init)
classificationKMEANSGAUSS= KMEANS(X,μ_init)
classificationHACGAUSS = HAC(X,3,Dmin)
S Curve
S1 = datasets.make_s_curve(n_samples=125, noise=.05)
S2 = datasets.make_s_curve(n_samples=125, noise=.05)
S = np.concatenate((S1[0],S2[0]-1.75))
X = np.zeros((1,250,2))
X[0,:,0] = S[:,0]
X[0,:,1] = S[:,2]
XS = X
classificationDBS = DBSCAN(X,0.5,10,euclidDist)
μ_init = np.array([[1,1] for i in range(2)])
μ_init[0][0] = -1
μ_init[1][1] = -1
Σ_init= np.array([np.identity(2)*0.1 for i in range(2)])
π_init = np.array([0.5,0.5])
classificationGMMS = GMM(X, μ_init, Σ_init, π_init)
classificationKMEANSS = KMEANS(X,μ_init)
classificationHACS = HAC(X,2,Dmin)
Circles
X = datasets.make_circles(n_samples=250, noise=.05,factor=0.5)
X = np.array([X[0]])
XCIRCLE = X
classificationDBCIRCLE= DBSCAN(X,0.2,5,euclidDist)
μ_init = np.array([[1,1] for i in range(2)])
μ_init[0][0] = -1
μ_init[1][1] = -1
Σ_init= np.array([np.identity(2)*0.1 for i in range(2)])
π_init = np.array([0.5,0.5])
classificationGMMCIRCLE = GMM(X, μ_init, Σ_init, π_init)
classificationKMEANSCIRCLE = KMEANS(X,μ_init)
classificationHACCIRCLE = HAC(X,2,Dmin)
Plots
fig, ax = plt.subplots(4,5,figsize=(20,16))
# OL FAITHFUL
for i in range(len(XOF[0])):
ax[0][0].scatter(XOF[0][i][0],XOF[0][i][1],color=colors[int(classificationDBOF[i])],alpha=0.5)
ax[1][0].scatter(XOF[0][i][0],XOF[0][i][1],color=colors[int(classificationGMMOF[i])+1],alpha=0.5)
ax[2][0].scatter(XOF[0][i][0],XOF[0][i][1],color=colors[int(classificationKMEANSOF[i])+1],alpha=0.5)
ax[3][0].scatter(XOF[0][i][0],XOF[0][i][1],color=colors[int(classificationHACOF[i])+1],alpha=0.5)
# MOONS
for i in range(len(XMOON[0])):
ax[0][1].scatter(XMOON[0][i][0],XMOON[0][i][1],color=colors[int(classificationDBMOON[i])],alpha=0.5)
ax[1][1].scatter(XMOON[0][i][0],XMOON[0][i][1],color=colors[int(classificationGMMMOON[i])+1],alpha=0.5)
ax[2][1].scatter(XMOON[0][i][0],XMOON[0][i][1],color=colors[int(classificationKMEANSMOON[i])+1],alpha=0.5)
ax[3][1].scatter(XMOON[0][i][0],XMOON[0][i][1],color=colors[int(classificationHACMOON[i])+1],alpha=0.5)
# GAUSS
for i in range(len(XGAUSS[0])):
ax[0][2].scatter(XGAUSS[0][i][0],XGAUSS[0][i][1],color=colors[int(classificationDBGAUSS[i])],alpha=0.5)
ax[1][2].scatter(XGAUSS[0][i][0],XGAUSS[0][i][1],color=colors[int(classificationGMMGAUSS[i])+1],alpha=0.5)
ax[2][2].scatter(XGAUSS[0][i][0],XGAUSS[0][i][1],color=colors[int(classificationKMEANSGAUSS[i])+1],alpha=0.5)
ax[3][2].scatter(XGAUSS[0][i][0],XGAUSS[0][i][1],color=colors[int(classificationHACGAUSS[i])+1],alpha=0.5)
# S
for i in range(len(XS[0])):
ax[0][3].scatter(XS[0][i][0],XS[0][i][1],color=colors[int(classificationDBS[i])],alpha=0.5)
ax[1][3].scatter(XS[0][i][0],XS[0][i][1],color=colors[int(classificationGMMS[i])+1],alpha=0.5)
ax[2][3].scatter(XS[0][i][0],XS[0][i][1],color=colors[int(classificationKMEANSS[i])+1],alpha=0.5)
ax[3][3].scatter(XS[0][i][0],XS[0][i][1],color=colors[int(classificationHACS[i])+1],alpha=0.5)
# CIRCLE
for i in range(len(XCIRCLE[0])):
ax[0][4].scatter(XCIRCLE[0][i][0],XCIRCLE[0][i][1],color=colors[int(classificationDBCIRCLE[i])],alpha=0.5)
ax[1][4].scatter(XCIRCLE[0][i][0],XCIRCLE[0][i][1],color=colors[int(classificationGMMCIRCLE[i])+1],alpha=0.5)
ax[2][4].scatter(XCIRCLE[0][i][0],XCIRCLE[0][i][1],color=colors[int(classificationKMEANSCIRCLE[i])+1],alpha=0.5)
ax[3][4].scatter(XCIRCLE[0][i][0],XCIRCLE[0][i][1],color=colors[int(classificationHACCIRCLE[i])+1],alpha=0.5)
# TITLES:
cols = ['Ol Faithful','Moons','3 Gaussians','Double S', 'Circles']
rows = ['DBSCAN','GMM','K Means','HAC']
for a, col in zip(ax[0], cols):
a.set_title(col,fontweight="bold")
for a, row in zip(ax[:,0], rows):
a.set_ylabel(row, rotation=90, size='large',fontweight="bold")
# Disable Ticks
for i in range(4):
for j in range(5):
ax[i][j].tick_params(
axis='x', # changes apply to the x-axis
which='both', # both major and minor ticks are affected
bottom=False, # ticks along the bottom edge are off
top=False, # ticks along the top edge are off
labelbottom=False) # labels along the bottom edge are off
ax[i][j].tick_params(
axis='y', # changes apply to the x-axis
which='both', # both major and minor ticks are affected
bottom=False, # ticks along the bottom edge are off
top=False, # ticks along the top edge are off
labelbottom=False) # labels along the bottom edge are off
fig.tight_layout()
plt.savefig("Clustering Algorithm Comparison")